A Plant Kingdom-Wide Structural Database of the CBP60 Protein Family.
2376 Transcripts
1996 Structures
1052 Genes
62 Species
2376 Transcripts
1996 Structures
1052 Genes
62 Species
The CBP60 protein family comprise a diverse set of proteins in plants (Zheng et al., 2021; Truman et al., 2013; Reddy et al., 2002). The best characterized members of this family are CBP60g and SARD1, which function as master transcription factors of plant immunity in the model species Arabidopsis thaliana (Sun et al., 2015; Truman and Glazebrook., 2012).
CBP60g and SARD1 are redundantly involved in pathogen-induced biosynthesis of central plant defence hormone salicylic acid (Wang et al., 2009; Zhang et al., 2010; Wang et al., 2011).
CBP60g and SARD1 also mediate the production of the major systemic immunity-inducing metabolite N-hydroxypipecolic acid (Sun et al., 2017; Sun et al., 2019).
Recently, it was reported that CBP60g and SARD1 serve as the primary, rate-limiting step controlling the temperature-vulnerability of the plant immune system (Kim et al., 2022).
Rational engineering of the plant CBP60 protein family could be a broadly applicable mechanism to safeguard the plant immune system and strengthen its resilience to a warming climate (Kim et al., 2022).
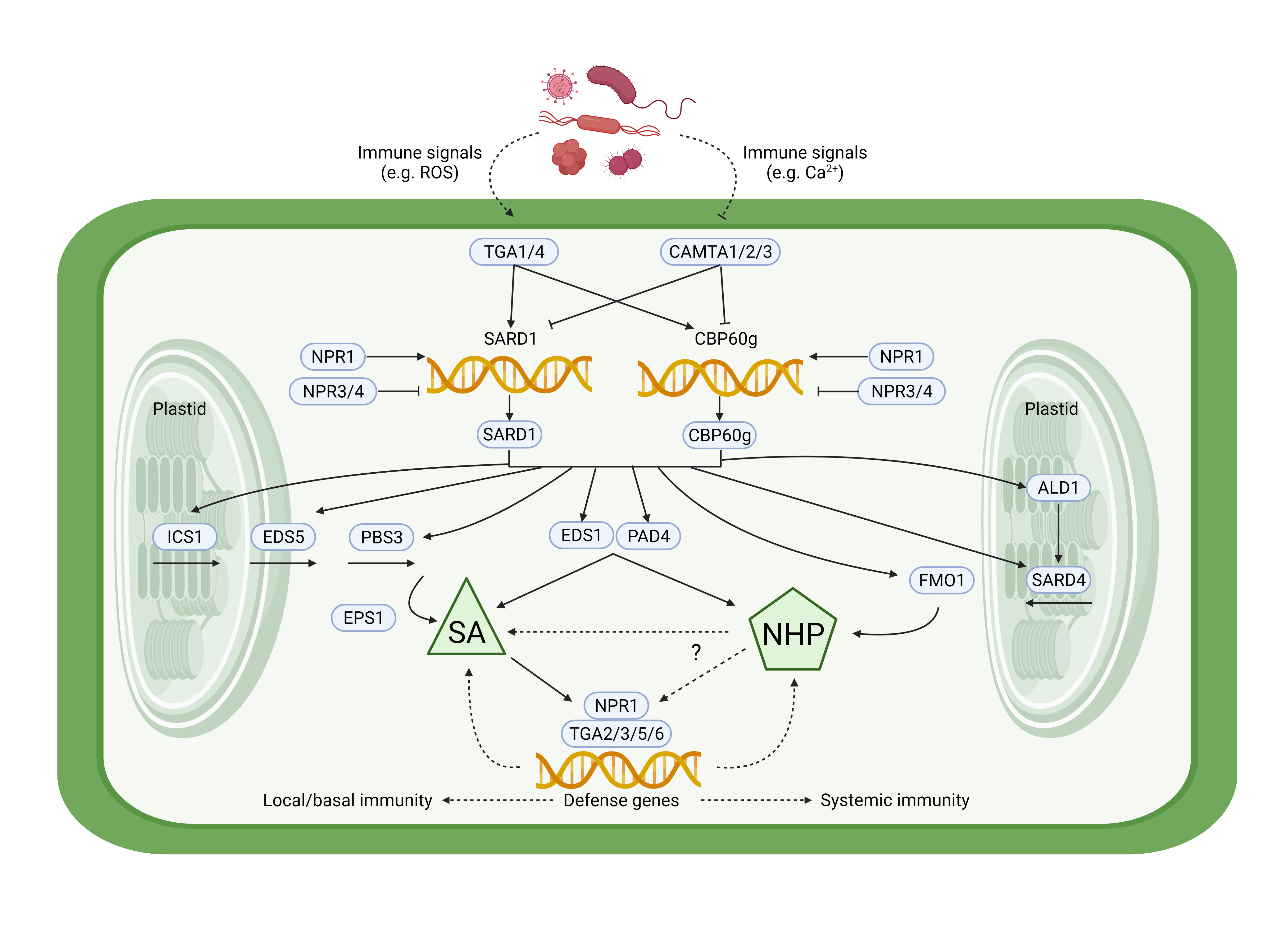
CBP60g and SARD1 as master transcription factors regulating basal and systemic immunity in plants. Shields et al., 2022; Figure 1.
The following plot is a 2D projection / cluster of all proteins within the database by their predicted structure. Proteins that appear closer together / have smaller euclidean distances between them can be interpreted as being more structurally similar. The primary metric used for clustering is the TM-Align score. For more information on how this projection was created please check out our paper. The TM-Align Cluster is available for download below, as well as through the programmatic API.
Download TM-Align Cluster Create Your OwnThe phylogenetic tree below was created within MEGA using the MUltiple Sequence Comparison by Log-Expectation (MUSCLE) alignment algorithm, as well as the FastTree phylogenetic tree creator. MUSCLE was used to create an alignment of all the AA sequences available within the database in the format of a FASTA file. This FASTA alignment was then exported and used by FastTree to produce the phylogenetic tree below in the Newick tree format. The visualization tool used is Phylocanvas. Both the FASTA alignment and the Newick tree are available for download below, as well as through the programmatic API.
75 University Avenue West, Waterloo, ON N2L 3C5
dcastroverde(at)wlu(dot)ca
519.884.0710 x3552